|
Welcome to the
CISN website
CISN: a software for confidence intervals of the
nucleotide substitution number
ˇ@
A major task in the exploration of an evolutionary process is to estimate the
substitution number per site of a protein or DNA sequence. The confidence interval
approach for estimating the substitution number can provide a more precise
estimation than the point estimation. Recently, useful confidence intervals have been
constructed for estimating the nucleotide substitution number. By applying the
confidence interval procedure, we first need to align sequences, and then apply the
procedure on the aligned sequences. Therefore, to develop a program which can
combine both techniques to provide a convenient tool for nucleotide substitution
number estimation is essential. In this study, we develop an R program to provide an
easy way for the nucleotide substitution number estimation which integrates the alignment procedure of Bioconductor software and R codes of the confidence interval
procedure. This integrated program can obtain 5 different confidence intervals for
the nucleotide substitution number by comparing two nucleotide sequences. Moreover,
it is likely that empirical information may exist in real application that the substitution
number may be within a specific range. In order to broaden the applicability of the
program, we associate the empirical information with the observed data in the
program to obtain more reliable confidence intervals, which can provide more precise
information for the nucleotide substitution number. ˇ@ |
ˇ» Evolutionary Models
ˇ´ JC69 model (Jukes and Cantor, 1969) (CISN-JC69)
ˇ´ K80 model (Kimura, 1980) (under investigation)
ˇ´ F81 model (Felsenstein, 1981) (under investigation)
ˇ´ HKY85 model (Hasegawa, Kishino and Yano, 1985) (under investigation)
ˇ´ T92 model (Tamura, 1992) (under investigation)
ˇ´ TN93 model (Tamura and Nei, 1993) (under investigation)
ˇ´ GTR: Generalised time-reversible (Tavaré, 1986) (under investigation)
ˇ» Reference
ˇ´ Wang, H. (2011). Confidence Intervals for the Substitution Number in the
Nucleotide Substitution Models. Molecular Phylogenetics Evolution, 60,
472-479.
ˇ´ Wang, H., Tzeng, Y.H. and Li, W.H. (2008). Improved variance estimators
for one- and two-parameter models of nucleotide substitution. Journal of
Theoretical Biology, 254, 164-167.
ˇ´ Wang, H. and Chen, W.S. CISN-JC69: an integrated program for confidence
intervals of the nucleotide substitution number for JC69 model. Submitted.
ˇ» Download the R-codes
| ˇ@ |
Download linkˇ@ |
File typeˇ@ |
File sizeˇ@ |
Updateˇ@ |
| R codesˇ@ |
CISN-JC69ˇ@ |
txtˇ@ |
9.95KBˇ@ |
2013.04.15ˇ@ |
The R project web for free download is
http://www.r-project.org.
Please ensure you already have installed R to run CISN codes.
ˇ» Run the code.
ˇ» R code user manual.
ˇ» A data Example
Data source:
ˇ´ J.P. Dumbacher, T.K. Pratt, and R.C. Fleischer, (2003). Phylogeny of the
owlet-nightjars (Aves: Aegothelidae) based on mitochondrial DNA sequence,
Molecular Phylogenetics Evolution, 29, 540-549.
ˇ´ Wang, H. and Hung, S.L. (2012). Phylogenetic tree selection by the adjusted
K-means approach. Journal of Applied Statistics, 39, 643-655.
The data set for the genome of the 11 owlet-nightjars.
| ˇ@ |
Download linkˇ@ |
File typeˇ@ |
File sizeˇ@ |
Updateˇ@ |
| Data file |
DATAˇ@ |
rar |
4.26KBˇ@ |
2012.12.11ˇ@ |
ˇ@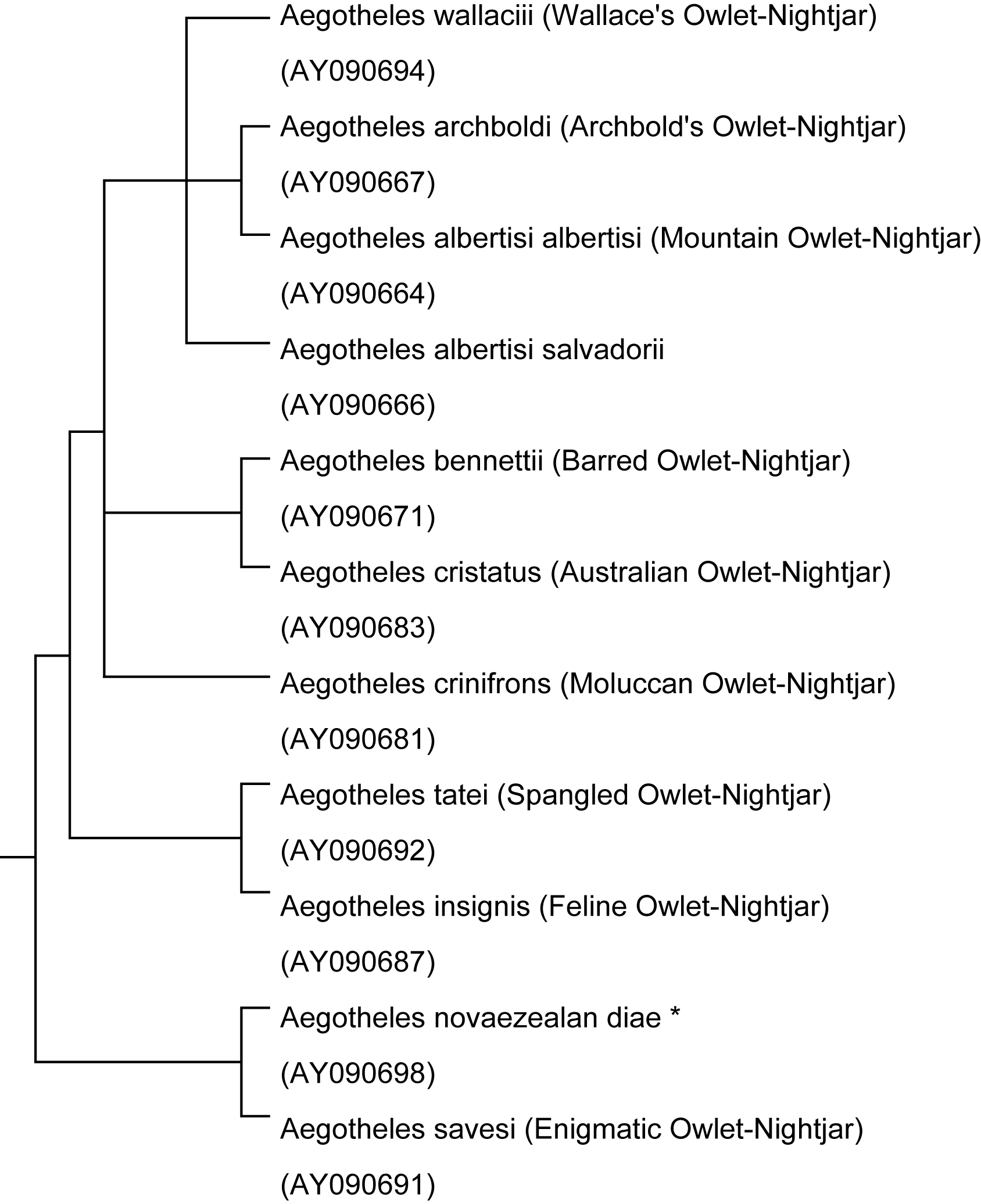
Figure. A simple form of the tree for the avian family Aegothelidae of the above data set.
ˇ» Contact
If you have any questions on CISN codes, please feel free to contact us at
wang@stat.nctu.edu.tw. We welcome your feedback and comments.
ˇ@
|